BCH5425 Molecular Biology and Biotechnology
Spring 1998
Dr. Michael Blaber
blaber@sb.fsu.edu
Lecture 8
DNA Supercoiling, cont., topoisomerases
A small circularly closed genome
The Simian Virus 40 (SV40) genome is a circular, closed, double stranded DNA genome. For the purposes of this discussion, it has 5300 bases. We expect that under physiological conditions the DNA will exhibit 10.6 base pairs per turn (i.e. one Twist = 10.6 bp/turn). In this case, with no Writhe, the Linking number would be:
Linking number = 5300 bp/(10.6 bp/turn) + 0
Linking number = 500 turns
i.e. we would expect 500 360° turns of the DNA strands over the length of the circular genome.
- This form (with 10.6 base pairs per turn) with no Writhe represents the "standard", or undistorted, DNA helix.
- This is also known as the "relaxed" form of DNA, and the duplex could physically be laid out flat on a surface because it needs no Writhe to achieve the preferred value of 10.6 basepairs per twist:
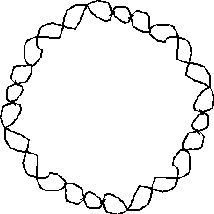
However, when the replication of SV40 is initially completed it is observed that there remains an open duplex region in the DNA:
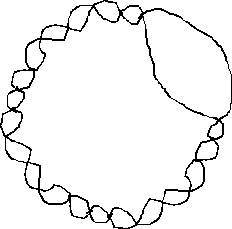
The result is that there are about 475 turns of the helix within the duplex DNA (i.e. the Linking number = 475).
- The DNA is said to be underwound.
- An open area is energetically unfavorable.
- The covalently closed molecule cannot adjust for this by increasing the Linking number
. That is, it cannot spontaneously break one or both strands of the duplex, introduce another 25 turns into the duplex (increase the Linking number by 25) and re-ligate the duplex.
The DNA has three choices:
- It can adjust the number of basepairs per turn throughout the molecule from a desired 10.6 bp/turn to 11.2 bp/turn (i.e. 5300 bp/475 turns). (NOTE: an increase in the number of basepairs per turn will decrease the twist value; underwound DNA has a greater number of basepairs per turn).
- The DNA can coil up into a "supercoil" topology and maintain the desired twist value (10.6) with the given linking number (475 in this case).
- The duplex can exist with a twist of 10.6 bp/turn for most of the structure, and then have a region with zero twist (not necessarily a melted duplex). This is quite unfavorable due to the geometry required of bond angles.
Thus for the 5300 bp SV40 genome, with a Linking number of 475, to maintain a value of 10.6 bp/twist, a total of 25 negative supercoils (Writhe=25) are needed:
475 = (5300/10.6) + Writhe
-25 = Writhe
That is, 25 negative supercoils (twenty five 180° turns of the DNA duplex, right handed as you look down the supercoiling).
Topoisomerases
The enzymes that control DNA topology are critical to DNA replication and transcription.
- As the replication fork opens up, the region of the duplex in front of the fork becomes overwound - i.e. it has fewer basepairs per turn.
- The linking number has not changed, but the length of DNA which contains all the turns is effectively shorter.
- To maintain 10.6 bp/turn in that region, the DNA will adopt positive supercoils.
For example, during the early stages of SV40 replication, the duplex around the origin of replication may initially melt (open up) a region of 750 bases. Since the Linkage number (500) is unchanged, it is effectively distributed over only:
5300 - 750 = 4550 bases.
Assuming no supercoiling has been introduced:
500 = 4550 basepairs / (X basepairs/twist) + 0
= 9.1 basepairs/twist
Thus, if no supercoiling is introduced, the DNA must adopt a conformation of 9.01 base pairs/twist of the helix within the region ahead of the replication fork.
- This is energetically unfavorable, and one option for the DNA is to adopt a supercoiled configuration to achieve 10.6 bp/twist:
500 = (4550/10.6) + Writhe
70.8 = Writhe
Thus, movement of the growing fork causes the DNA to adopt positive supercoils.
In this case the DNA has adopted 70.8 left handed supercoils (180° each).
Twist (=basepairs * [twist/basepair]) and Writhe are both real numbers.
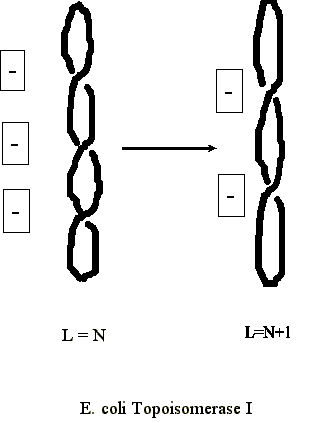
Type I Topoisomerase
- Type I topoisomerases cut one strand of the DNA (i.e. it "nicks" the DNA duplex).
- The 5' phosphate of the nicked strand is covalently attached to a tyrosine in the protein.
- The 3' end of the nick then passes once through the duplex.
- The nick is then resealed, and the linkage number is change by a value of +1.
- This can therefore result in the removal of a single negative supercoil.
In E. coli, type I topoisomerase can only relieve negatively supercoiled DNA (negative supercoiling is the end result of newly replicated DNA genome). In eukaryotes, type I topoisomerase can also relieve positively supercoiled DNA.
The net result of E. coli topoI can be diagrammed as follows:
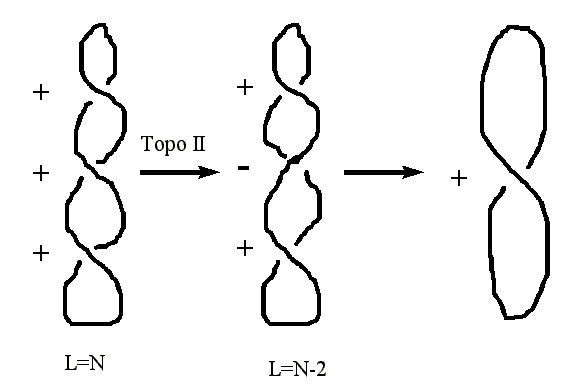
Type II Topoisomerases
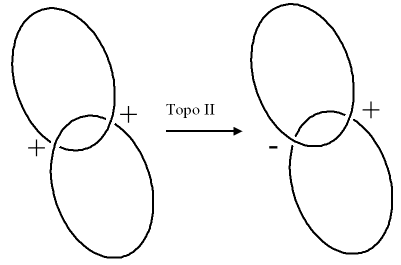
- Type II topoisomerases actually cleave the duplex DNA in changing the linkage number.
- Type II topoisomerases can convert a single positive supercoil into a negative supercoil.
- Thus the linkage number is reduced by two (-2) in a single step.
- Type II topoisomerases are involved in both decatenation of daughter chromosomes, and relieving the positive supercoiling ahead of the replication fork.
- E. coli
DNA gyrase is an example of a type II topoisomerase.
1998 Dr. Michael Blaber

